Crispr-Cas9 (Apo form)
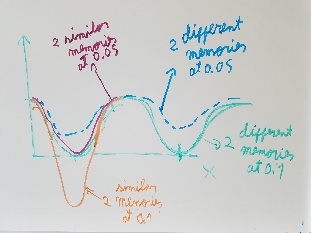
- The framgent memory force in this simulation was too big (0.2), I'm repeating the simulation with a smaller force (0.01). I expect that the energy of the local interactions (RAMA+HELIX+F.M.) should contribute around 2/3 of the energy of the non-local interactions (DSSP,PAP,WATER)
...
Use fragment memory force for the parts of the protein that are flexible. (Running simulations with dual memory, with a MF of 0.05. Should be equivalent to a simulation of SMF 0.1 but with two 0.05 wells around the region where the conformation is diferent. Running with and without DNA)