...
Convert the log file generated by our MCMC _SEQ sampling method to be readable by Tracer.
PhyloNet Version: 3.6.3
Usage
|
|---|
| Settings | ||
-cl chainLength | The length of the MCMC chain. Set to the same value used in MCMC_SEQ. | required |
| -bl burnInLength | The number of iterations in burn-in period. Set to the same value used in MCMC_SEQ. | required |
-sf sampleFrequency | The sample frequency. Set to the same value used in MCMC_SEQ. | required |
| -outfile filename | The name of tracer log file you want to generate. Can contain path. | required |
| -truenet networkString | The network you want to summarize its parameters. | required |
| MC3 Settings | ||
| -mc3 temperatureList | The list of temperatures for the Metropolis-coupled MCMC chains. For example, -mc3 (2.0, 3.0)indicates two hot chains with temperatures 2.0 and 3.0 respectively will be run along with the cold chain with temperature 1.0. By default only the cold chain will be run. Note that
| optional |
| Inference Settings | ||
| -mr maxReticulation | The maximum number of reticulation nodes in the sampled phylogenetic networks. The default value is 4. | optional |
| -taxa taxaList | The taxa used for inference. For example, -taxa (a,b,c) | required |
| -tm taxonMap | Gene tree / species tree taxa association. By default, it is assumed that only one individual is sampled per species in gene trees. This option allows multiple alleles to be sampled. For example, the gene tree is (((a1,a2),(b1,b2)),c); and the species tree is ((a,b),c);, the command is -tm <a:a1,a2; b:b1,b2;c:c>. Note that the taxa association should cover all species, e.g. -tm <a:a1,a2; b:b1,b2> is incorrect because c:c is dropped out. | optional |
| -fixtheta theta | Fix the population mutation rates associated with all branches of the phylogenetic network to this given value (theta). By default, we estimate a constant population size across all branches. | optional |
| -varytheta | The population mutation rates across all branches may be different when estimating them. By default, we estimate a constant population size across all branches. | optional |
| -esptheta | Estimate the mean value of prior of population mutation rates. | optional |
| Prior Settings | ||
| -pp poissonParam | The Poisson parameter in the prior on the number of reticulation nodes. The default value is 1.0. | optional |
| -dd | Disable the prior on the diameters of hybridizations. By default this prior on is exp(10). | optional |
| -ee | Enable the Exponential(10) prior on the divergence times of nodes in the phylogenetic network. By default we use Uniform prior. | optional |
| Starting State Settings | ||
| -snet | Specify the starting network. The input network should be ultrametric with divergence times in units of expected number of mutations per site, inheritance probabilities and population sizes in units of population mutation rate (optional). See example below. The default starting network is the MDC trees given starting gene trees. | optional |
| -ptheta startingThetaPrior | Specify the mean value of prior of population mutation rate (startingThetaPrior). The default value is 0.036. If -esptheta is used, startingThetaPrior will be treated as the starting value, otherwise startingThetaPrior will be treated as the fixed mean value of prior of population mutation rates. | optional |
Data related settings | ||
| -diploid | Specify whether sequence sampled from diploids. | optional |
| -dominant dominantMarker | Specify which marker is dominant if the data is dominant. Either be '0' or '1'. | optional |
| -op | Specify whether or not to ignore all monomorphic sites. If this option is used, the data will be treated as containing only polymorphic sites. | optional |
Example
#NEXUS
Begin data;
Dimensions ntax=5 nchar=10;
Format datatype=dna symbols="012" missing=? gap=-;
Matrix
b 1100100110
c1 1010100011
c2 1001110100
d 1000011110
;End;
BEGIN PHYLONET;
MCMC_BiMarkers -cl 500000 -bl 200000 -sf 500 -diploid -dominant 1 -op -varytheta -pp 2.0 -ee 2.0 -mr 1 -pl 4 -esptheta -ptheta 0.3
-sd 12345678
-taxa (a,b,c1,c2,d)
-tm <A:a; B:b;C:c1,c2;D:d>;
END;
Example
If we run MCMCseq_example1.nex , which is one of the example in the description of command MCMC_SEQ, we will get following output. Suppose it is printed into file example_mcmcseq_1.out.
Then the content of example_mcmcseq_1.out looks like:
MCMC_SEQ -cl 50000 -bl 10000 -sgt (gt0, gt1) -snet (net1) -sps 0.04 -pre 20 Overall MAP = -2367.294357178221 Total elapsed time : 23.77700 s |
|---|
Suppose we want to analyze the parameters of sampled top network using Tracer, we can write such a NEXUS file. Note that the "truenet" string is copied from the "Rank = 0" network. And remember to put the log file into SETS section.
#NEXUS BEGIN SETS; BEGIN PHYLONET; |
|---|
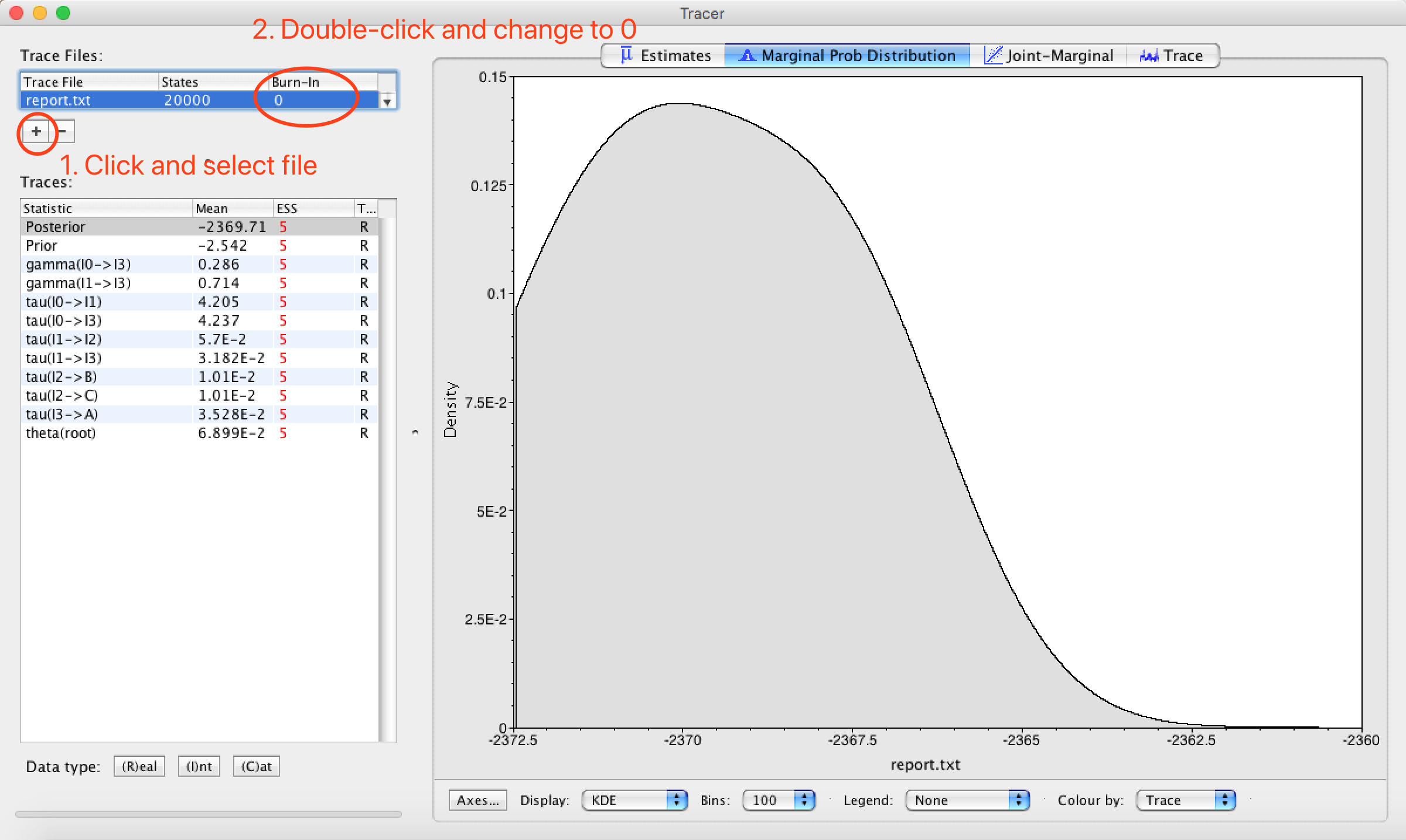
Run this NEXUS file using PhyloNet. PhyloNet will print a string showing how nodes are labeled, and report.txt is generated, which is readable by Tracer.
Following is what we will see after import report.txt into Tracer. In the column of Statistic, gamma represents inheritance probability, tau represents branch length, and theta represents population mutation rate.
Note that an empty line should be left after "Matrix".
This command will run MCMC chain of 500000 iterations with 200000 burn-in iterations, and one sample will be collected every 500 iterations. The taxa are diploids and 1 is the dominant marker. Only polymorphic sites will be used. We will estimate population mutation rates for every branches, and they may be different. A Poisson prior of 2.0 will be adopted, and a Exponential(2.0) prior will be adopted. The number of reticulation nodes is limited to 1. We will sample the mean value of prior of population mutation rates, and the starting value of 0.3 is given. We use the random seed of 12345678. In the end, we indicate the mapping from taxa to species.