...
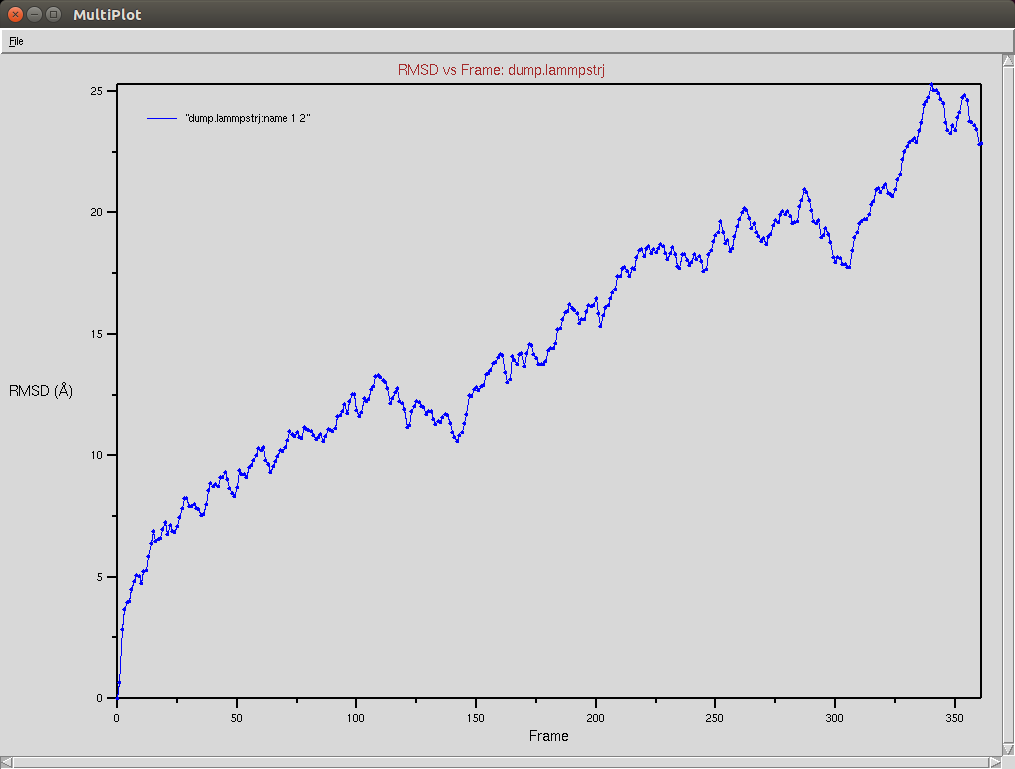
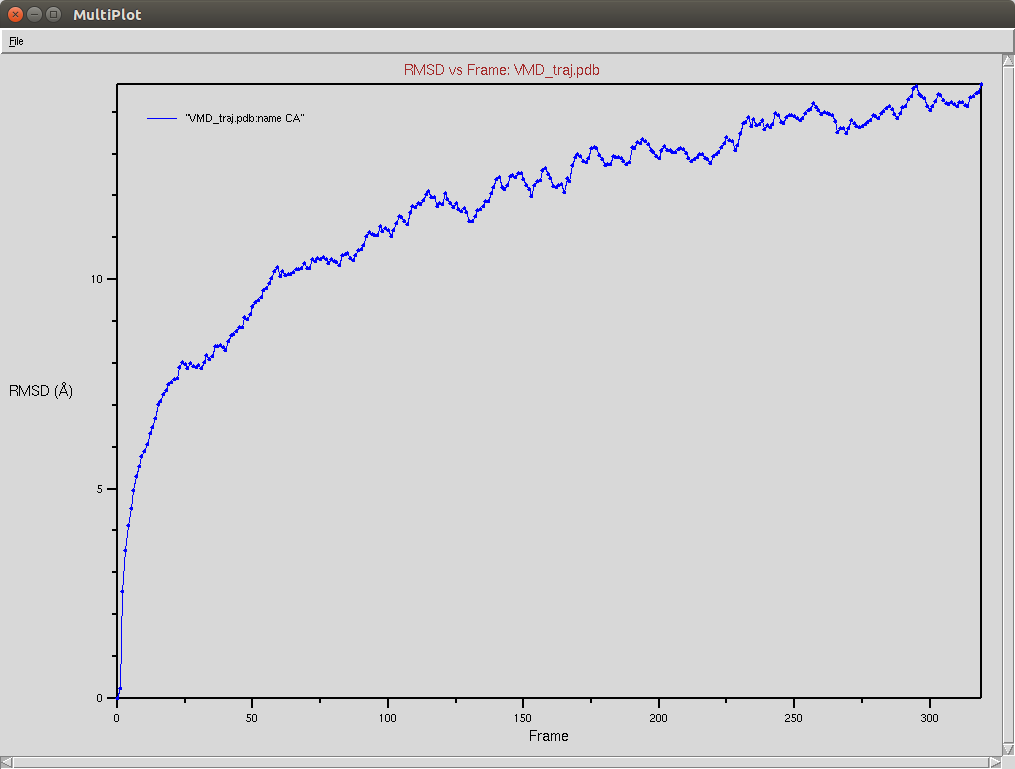
Nucleic RMSD
CRISPR-Cas9+DNA+RNA
The Cas9-DNA-RNA system was simulated using a SMF of 0.5
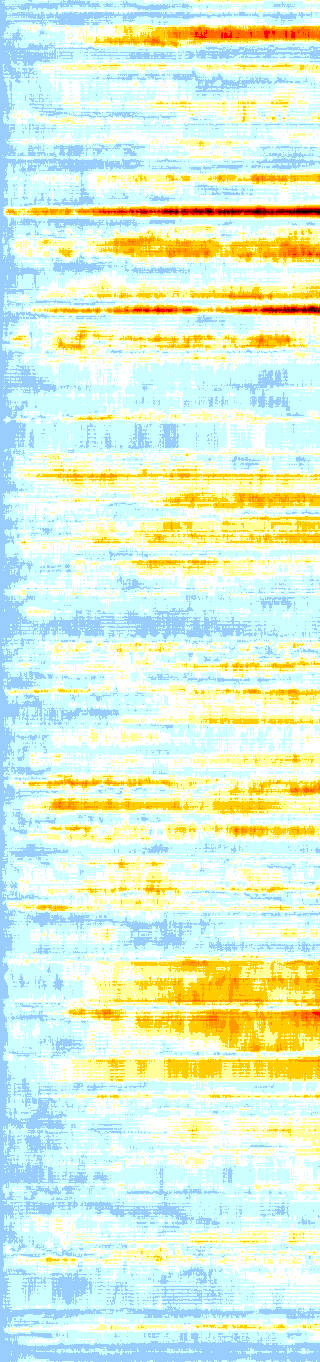
Protein heatmap
Some technical constraints
It is still difficult to mix two nucleic chain together (like DNA and RNA) because merge.py only accepts two files. (Working on thisSolved by merging the DNA and RNA before merging it with the protein)
The simulation seems to be running slowly because there are too many neighbours on the program, it seems that for the central atoms the number of neighbors is more than 2000. This may be caused because of the electrostatic potential cutoff.
AWSEM-3SPN.2 doesnt doesn't run with the new version (it almost runs, but atoms from AWSEM dissapear, with the same configuration that worked on the old version)
The premerge script also needs some modifications if I want to simulate the RNA in A-form. Currently it is impossible to simulate RNA with the A-form forcefield, but the 3SPN.2C seems to keep the RNA structure.
It is impossible to restart the simulations, since the read_restart doesn't work with 3SPN. (A possible fix may be on the new lammps version with read_restart remap)