| Table of Contents |
|---|
1. Introduction
PhyloNet is a tool designed mainly for analyzing, reconstructing, and evaluating reticulate (or non-treelike) evolutionary relationships, generally known as phylogenetic networks. Various methods that we have developed make use of techniques and tools from the domain of phylogenetic trees, and hence the PhyloNet package includes several tools for phylogenetic tree analysis. PhyloNet is released under the GNU General Public License. For the full license, see the file GPL.txt included with this distribution.
1.1 Contributors
PhyloNet is designed, implemented, and maintained by Rice's BioInformatics Group, which is lead by Professor Luay Nakhleh (nakhleh@cs.rice.edu). For more details related to this group please visit http://bioinfo.cs.rice.edu.
This tutorial is based on the book chapter: Practical Aspects of Phylogenetic Network Analysis Using PhyloNet
2. Installation
System Requirements
In order to run the PhyloNet toolkit, you must have Java 1.7.0 or later installed on your system. All references to the java command assume that Java 1.7 is being used.
...
Place the jar file in the desired installation directory. The remainder of this document assumes that it is located in $PHYLONET PATH/jar. Installation is now complete. In order to run PhyloNet, you must execute the file PhyloNet_X.Y.Z.jar, as described in the next section.
3. Basic Usage
The PhyloNet tool is executed by typing the following command into your console:
...
Where script.nex is the NEXUS file containing the commands to be executed.
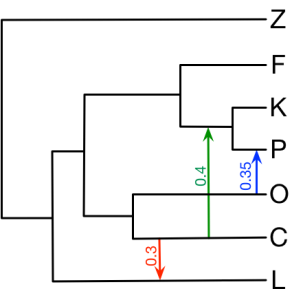
4. Illustrating the Various Inference Methods in PhyloNet
Here we provide all the input nexus files in section 4 of the book chapter. The figure below is the true network.
4.1 Minimize deep coalescent Inference
This section corresponds to section 4.1 of the book chapter.
4.1.1 MDC Inference using true gene tree topologies
| Input nexus file | Maximum number of reticulations |
|---|---|
| InferNetwork_MP_pl8_0_true.nex | 0 |
| InferNetwork_MP_pl8_1_true.nex | 1 |
| InferNetwork_MP_pl8_2_true.nex | 2 |
| InferNetwork_MP_pl8_3_true.nex | 3 |
| InferNetwork_MP_pl8_4_true.nex | 4 |
Corresponding results: Fig 4 in the book chapter.
4.1.2 MDC Inference using gene tree topologies estimated by IQTREE
| Input nexus file | Maximum number of reticulations |
|---|---|
| InferNetwork_MP_pl8_0_false.nex | 0 |
| InferNetwork_MP_pl8_1_false.nex | 1 |
| InferNetwork_MP_pl8_2_false.nex | 2 |
| InferNetwork_MP_pl8_3_false.nex | 3 |
| InferNetwork_MP_pl8_4_false.nex | 4 |
Corresponding results: Fig 5 in the book chapter.
4.2 Maximum Likelihood Inference
This section corresponds to section 4.2 of the book chapter.
4.2.1 ML Inference using true gene tree topologies
| Input nexus file | Maximum number of reticulations |
|---|---|
| InferNetwork_ML_pl8_0_true.nex | 0 |
| InferNetwork_ML_pl8_1_true.nex | 1 |
| InferNetwork_ML_pl8_2_true.nex | 2 |
| InferNetwork_ML_pl8_3_true.nex | 3 |
| InferNetwork_ML_pl8_4_true.nex | 4 |
Corresponding results: Fig 6 in the book chapter.
4.2.2 ML Inference using gene tree topologies estimated by IQTREE
| Input nexus file | Maximum number of reticulations |
|---|---|
| InferNetwork_ML_pl8_0_false.nex | 0 |
| InferNetwork_ML_pl8_1_false.nex | 1 |
| InferNetwork_ML_pl8_2_false.nex | 2 |
| InferNetwork_ML_pl8_3_false.nex | 3 |
| InferNetwork_ML_pl8_4_false.nex | 4 |
Corresponding results: Fig 6 in the book chapter.
4.2.3 ML Inference using true gene tree topologies and branch lengths
| Input nexus file | Maximum number of reticulations |
|---|---|
| InferNetwork_ML_bl_pl8_0_true.nex | 0 |
| InferNetwork_ML_bl_pl8_1_true.nex | 1 |
| InferNetwork_ML_bl_pl8_2_true.nex | 2 |
| InferNetwork_ML_bl_pl8_3_true.nex | 3 |
| InferNetwork_ML_bl_pl8_4_true.nex | 4 |
Corresponding results: Fig 7 in the book chapter.
4.2.4 ML Inference using gene tree topologies and branch lengths estimated by IQTREE
| Input nexus file | Maximum number of reticulations |
|---|---|
| InferNetwork_ML_bl_pl8_0_false.nex | 0 |
| InferNetwork_ML_bl_pl8_1_false.nex | 1 |
| InferNetwork_ML_bl_pl8_2_false.nex | 2 |
| InferNetwork_ML_bl_pl8_3_false.nex | 3 |
| InferNetwork_ML_bl_pl8_4_false.nex | 4 |
Corresponding results: Fig 9 in the book chapter.
4.3 Maximum Pseudo-likelihood Inference
This section corresponds to section 4.3 of the book chapter.
4.3.1 MPL Inference using true gene tree topologies
| Input nexus file | Maximum number of reticulations |
|---|---|
| InferNetwork_MPL_pl8_0_true.nex | 0 |
| InferNetwork_MPL_pl8_1_true.nex | 1 |
| InferNetwork_MPL_pl8_2_true.nex | 2 |
| InferNetwork_MPL_pl8_3_true.nex | 3 |
| InferNetwork_MPL_pl8_4_true.nex | 4 |
Corresponding results: Fig 10 in the book chapter.
4.3.2 MPL Inference using gene tree topologies estimated by IQTREE
| Input nexus file | Maximum number of reticulations |
|---|---|
| InferNetwork_MPL_pl8_0_false.nex | 0 |
| InferNetwork_MPL_pl8_1_false.nex | 1 |
| InferNetwork_MPL_pl8_2_false.nex | 2 |
| InferNetwork_MPL_pl8_3_false.nex | 3 |
| InferNetwork_MPL_pl8_4_false.nex | 4 |
Corresponding results: Fig 11 in the book chapter.
4.3.3 MPL Inference using bi-allelic marker data
| Input nexus file | Maximum number of reticulations |
|---|---|
| 0 | |
| 1 | |
| 2 | |
| 3 | |
| 4 |
Corresponding results: Fig 12 in the book chapter.
4.4 Bayesian Inference
4.4.1 MCMC_SEQ: Bayesian inference on the sequence alignment data
| Input nexus file | Maximum number of reticulations |
|---|---|
| 0 | |
| 1 | |
| 2 | |
| 3 | |
| 4 |
Corresponding results: Fig 13 in the book chapter.
4.4.2 MCMC_GT: Bayesian inference on gene tree topologies
4.4.2.1 MCMC_GT sampling using true gene tree topologies
| Input nexus file | Maximum number of reticulations |
|---|---|
| MCMC_GT_pl8_0_true.nex | 0 |
| MCMC_GT_pl8_1_true.nex | 1 |
| MCMC_GT_pl8_2_true.nex | 2 |
| MCMC_GT_pl8_3_true.nex | 3 |
| MCMC_GT_pl8_4_true.nex | 4 |
4.4.2.2 MCMC_GT sampling using gene tree topologies estimated by IQTREE
| Input nexus file | Maximum number of reticulations |
|---|---|
| MCMC_GT_pl8_0_false.nex | 0 |
| MCMC_GT_pl8_1_false.nex | 1 |
| MCMC_GT_pl8_2_false.nex | 2 |
| MCMC_GT_pl8_3_false.nex | 3 |
| MCMC_GT_pl8_4_false.nex | 4 |
4.4.3 MCMC_BiMarkers: Bayesian inference on the bi-allelic markers
| Input nexus file | Maximum number of reticulations |
|---|---|
| 0 | |
| 1 | |
| 2 | |
| 3 | |
| 4 |
...