...
Where $PHYLONET_PATH is the directory of jar file PhyloNet_X.Y.Z.jar, and script.nex is the NEXUS file containing the commands to be executed.
Scoring a candidate species phylogeny
- Parsimonious:
- Scoring a species tree (ILS): DeepCoalCount_tree
- Scoring a species tree/network (ILS+introgression): DeepCoalCount_network
- Probabilistic:
- Scoring a species tree/network (ILS+introgression): CalGTProb
Inferring a species phylogeny
- Maximum parsimony:
- Inferring a species tree (ILS): Infer_ST_MDC, Infer_ST_MDC_UR
- Inferring a species tree/network (ILS+introgression): InferNetwork_MP
- Maximum likelihood:
- Inferring a species tree/network (ILS+introgression): InferNetwork_ML
- inferring a species tree/network (ILS+introgression) with cross-validation: InferNetwork_ML_CV
- Maximum pseudo-likelihood:
- Inferring a species tree/network (ILS+introgression): InferNetwork_MPL
- Bayesian:
- Inferring a species tree/network (ILS+introgression): MCMC_GT
- Inferring a species tree/network (ILS+introgression) from multilocus data: MCMC_SEQ
- Inferring a species tree/network (ILS+introgression) from bi-allelic marker data: MCMC_BiMarkers
- Larger data sets:
- Inferring a species tree/network (ILS+introgression) by divide-and-conquer: NetMerger
- Inferring a species tree/network (ILS+introgression) while fixing the start tree topology: InferNetwork_MPL (-fs) and InferNetwork_MP (-fs)
Comparing and Summarizing
- Summarizing networks: SummarizeNetworks
For other tools in PhyloNet, please see here for a full list.
4. Illustrating the Various Inference Methods in PhyloNet
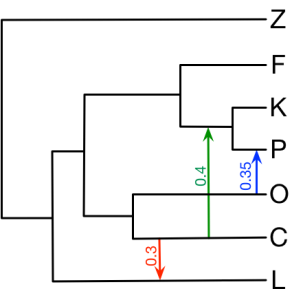
Here we provide all the input NEXUS files in section 4 of the book chapter. The figure below is the true network we would like to infer. You can find detailed usage of all the commands below on https://wiki.rice.edu/confluence/display/PHYLONET/List+of+PhyloNet+Commands .
4.1 Minimize deep coalescent Inference
...
This inference ran out of 192 CPU hours.
5. Visualizing a Phylogenetic Network
Phylogenetic network in Rich Newick string can be visualized in Dendroscope or icytree. The former needs downloading, and the latter is online. However, neither can recognize inheritance probabilities (branch lengths are fine). You have to remove those probabilities manually from the Rich Newick string, or use option "-di" so that PhyloNet returns the network that Dendroscope takes directly. For example, the following network is what PhyloNet returns when the input NEXUS file InferNetwork_MP is used:
| Code Block |
|---|
((((((Skud,(Sbay)#H1:::0.36082474226804123),((Spar,Scer),Smik)),#H1:::0.6391752577319587),Scas),Sklu),Calb); |
In order to visualize this phylogenetic network in Dendroscope, please remove the inheritance probabilities like follows, and Dendroscope will be able to read it. Or you can use option "-di".
| Code Block |
|---|
((((((Skud,(Sbay)#H1),((Spar,Scer),Smik)),#H1),Scas),Sklu),Calb); |