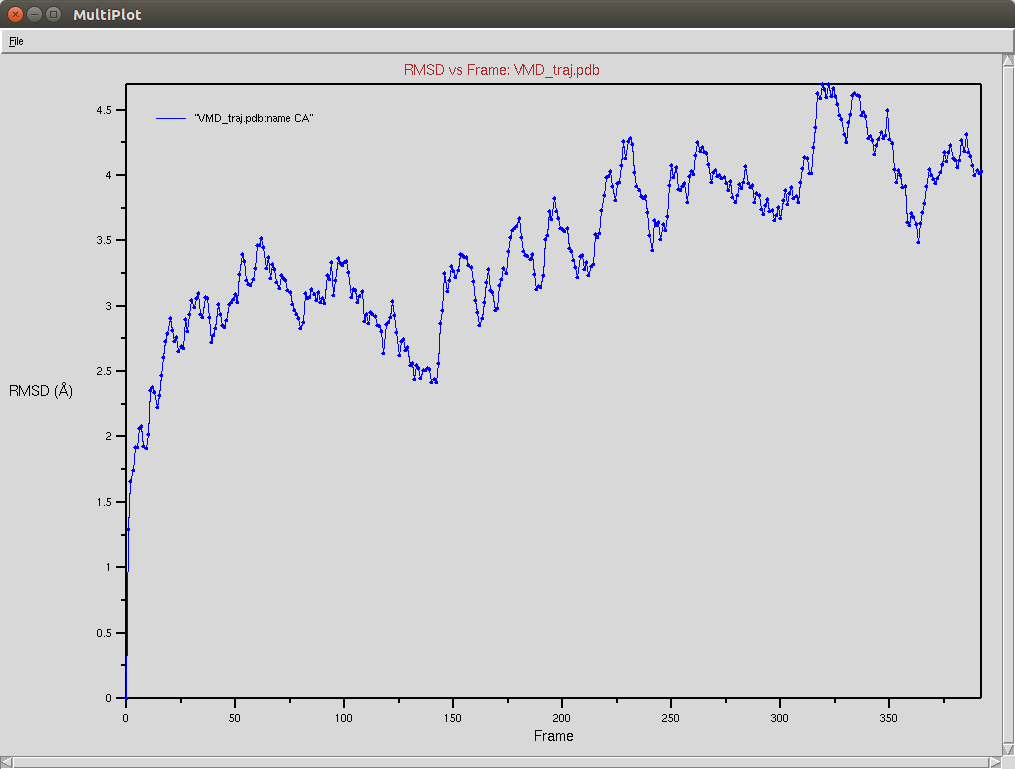
Crispr-Cas9 (Apo form)
- The framgent memory force in this simulation was too big (0.2), I'm repeating the simulation with a smaller force (0.01). I expect that the energy of the local interactions (RAMA+HELIX+F.M.) should contribute around 2/3 of the energy of the non-local interactions (DSSP,PAP,WATER)
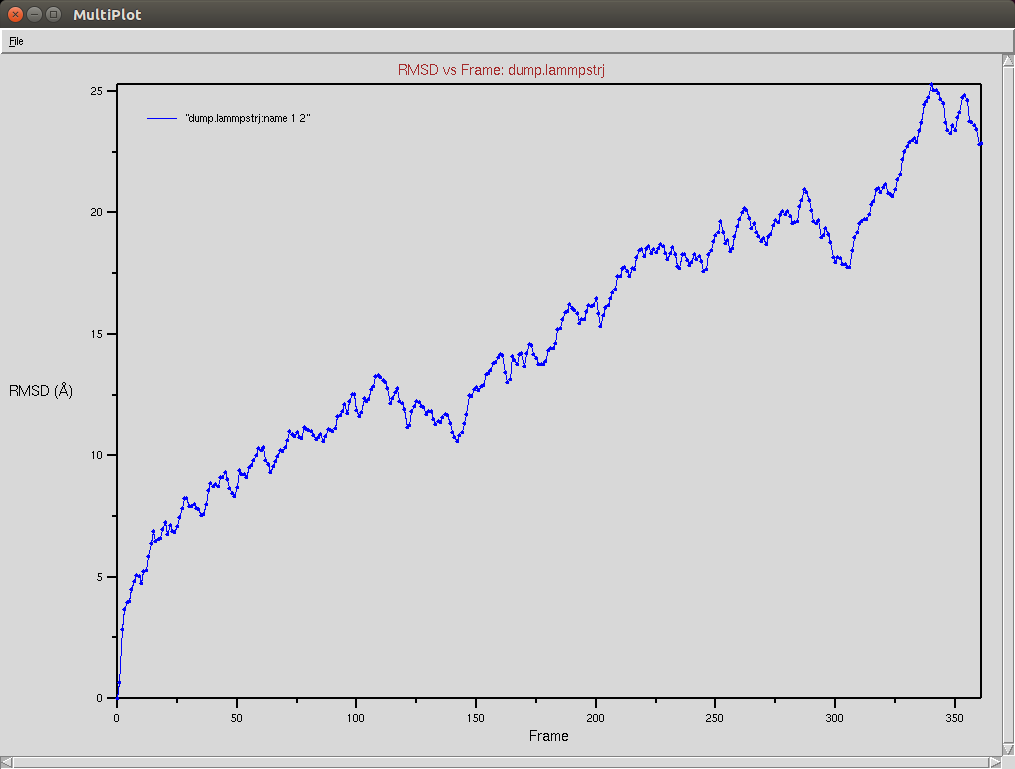
Protein RMSD
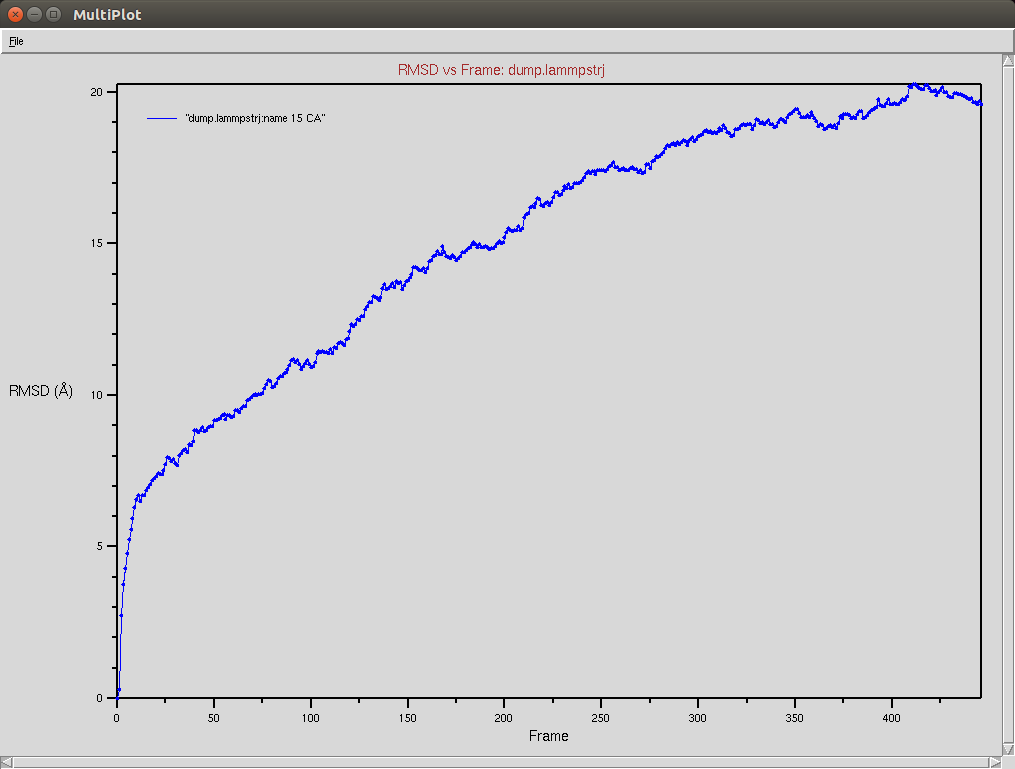
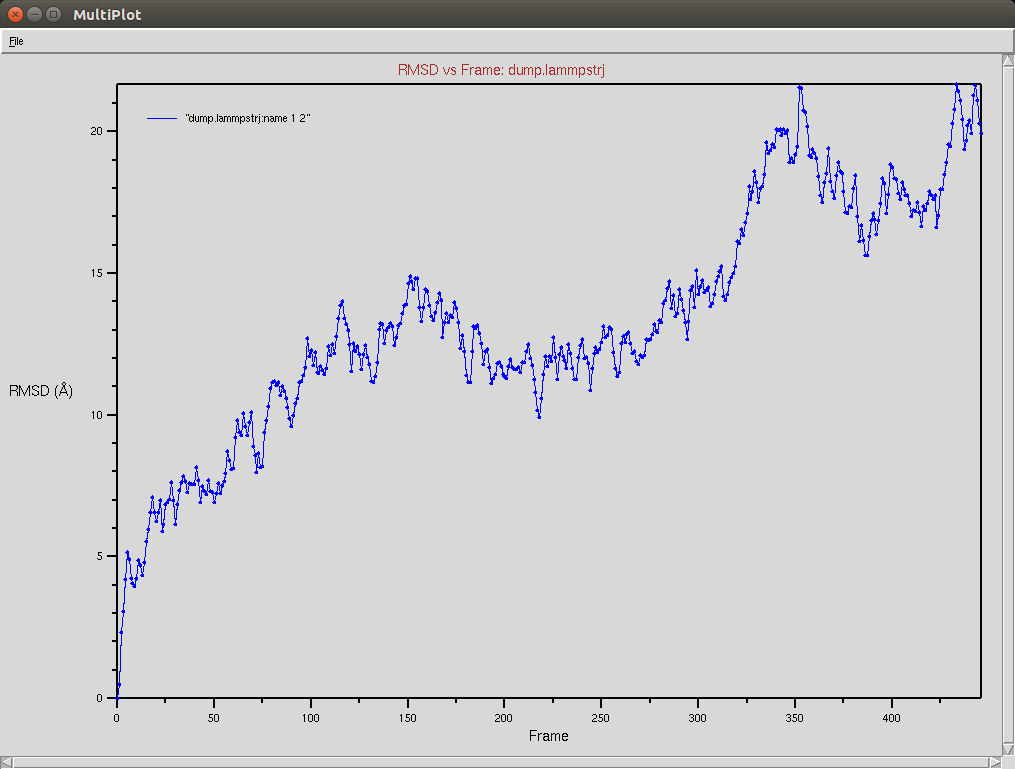
Crispr-Cas9 + DNA
Due to a mistake I didn't update the neighbor list, so it is possible to see an overlap between two domains (grey and red). The single memory force was turned off in this simulation.
Protein RMSD
Nucleic RMSD
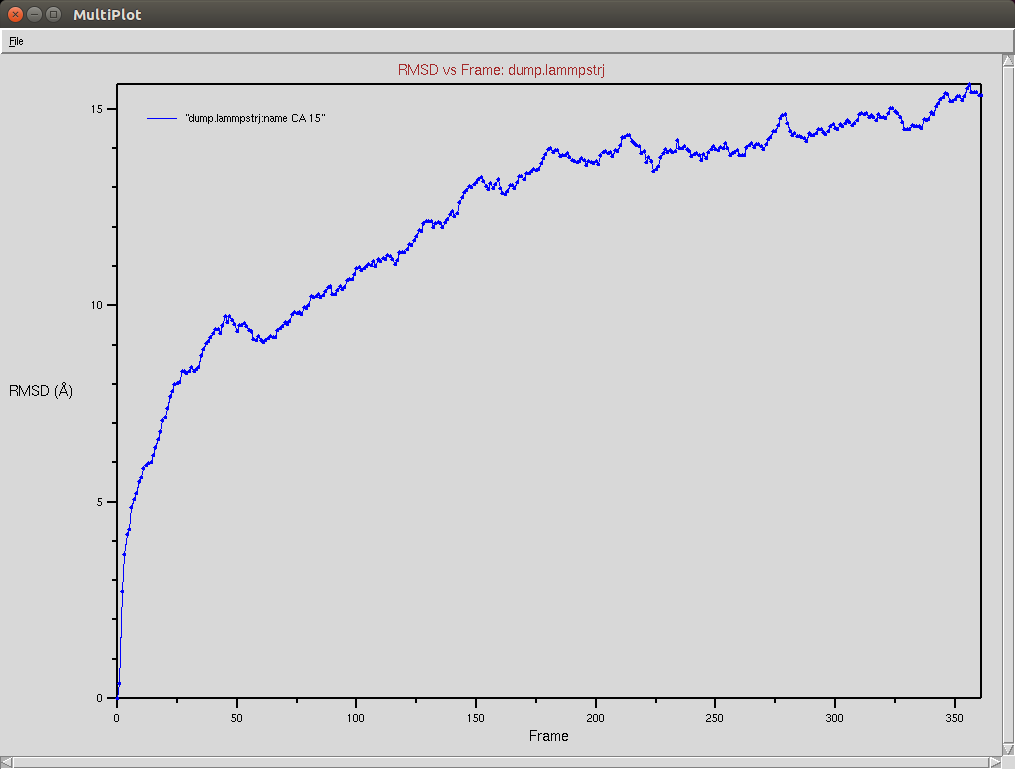
Crispr-Cas9 + RNA
Due to a mistake I didn't update the neighbor list, so it is possible to see an overlap between two domains (grey and red). The single memory force was turned off in this simulation.
Protein RMSD
Nucleic RMSD