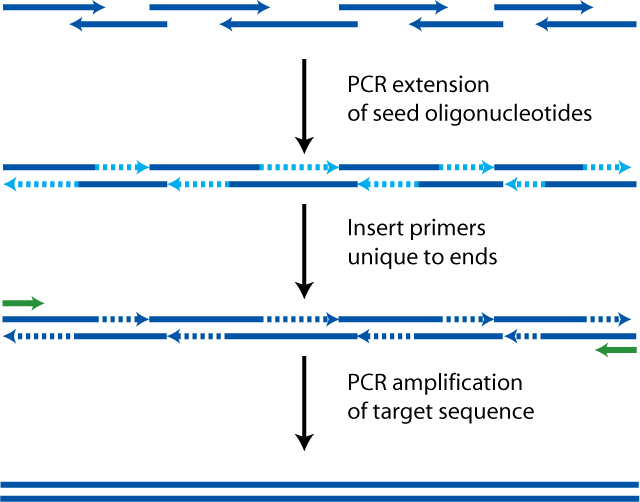
Polymerase chain assembly (PCA), also known as polymerase cycling assembly or PCR assembly, is useful in the de novo (template-less) generation of shorter linear DNA parts (<500 bp) by extending many partially-overlapping oligonucleotide primers off one another in a PCR to generate the full linear product. While the smallest parts are more cost-effectively generated by annealing pairs of oligonucleotides, PCA can be more efficient for longer small parts, as the polymerase in PCR synthesizes DNA segments between successive primers/amplicons on each strand using the opposite strand's primers/amplicons as templates, DNA which needn't be purchased pre-synthesized as oligos as when making oligo annealing products, for which every nucleotide of both strands must be purchased in oligos. Oligo annealing products, however, can be designed to intrinsically have cohesive "sticky" ends / overhangs, allowing them to omit the DNA encoding restriction sites for downstream assemblies, saving some cost compared to PCA, which just as any PCR product, requires including flanking restriction sites if using in restriction-based assembly such as Golden Gate.
For cost-minimization, compare the following:
- Cost of oligo-anneal product =
($/nt) × 2 × (lengthpart + lengthoverhang)- For typical part assembly, "part length" includes 2 flanking latent BsaI overhangs, but not the one ssDNA PaqCI overhang per strand.
Thus, for a pure, part-only sequence: ($/nt) × 2 × (12 nt + lengthpart-only)
- For typical part assembly, "part length" includes 2 flanking latent BsaI overhangs, but not the one ssDNA PaqCI overhang per strand.
- Cost of PCA product = ($/nt) × Σ(all PCA primer-lengths), where primers are designed in Primerize with any necessary terminal restriction sites and end-spacers.
- For typical part assembly, part-only sequence can be input into Primerize, primer lengths summed, and 34 nt added for the two 17 nt part adapters in the outermost primers.
Use the online Primerize software, following their tutorial.
- Make sure "check for T7 promoter" is unchecked, as this option adds a T7 promoter sequence to the design.
- Ensure any restriction sites (e.g. Golden Gate sites) or Gibson homologies are included at the ends as with any PCR product to be used in a later assembly.
- Primers are best kept ≤60 nt, as per-nucleotide price and synthesis/delivery time typically increases for >60 nt primers, as well as synthesis error rate.
- The PCA reaction protocol can be followed as described but with the higher-fidelity Q5 polymerase and buffer, and the 50 µL reaction size can be scaled down (along with components) to 20 µL with sufficient yield. The reaction is essentially a PCR containing all the designed primers but with the outer two primers at 100× the concentration of the inner primers.
- For cases where mispriming warnings are shown, consider the stepwise "multiple-round" PCA methods described, wherein two or more partially overlapping subfragments are first separately generated in "sub-pools", purified, and combined in a final PCA to make the full product. In the case Strategy 1: '2-Round' method does not work or is not predicted to due to the nature of the potential mispriming, the Strategy 2: 'Couple' reacts only pairs of primers first to make fragments for the final PCA.
PCA Primer Mix
- Final PCA rxn: 4 µM outer primers, 0.04 µM inner primers
- 10× primer mix: 40 µM outer primers, 0.4 µM inner primers
For 20 µL of 10× PCA primer mix:
- In one tube, make a 2 µM dilution mix of inner primers:
Combine 1 µL of each 100 µM inner primer + water in a final 50 µL. - In another tube, for 20 µL of 10× primer mix, combine:
- 8 µL 100 µM outer primer 1
- 8 µL 100 µM outer primer 2
- 4 µL 2 µM inner primer mix
- Use this 10× mix as 1/10 of a PCA, e.g. 2.5 µL in a 25 µL rxn.